Embedded CSE (eCSE) support
Through a series of regular calls, Embedded CSE (eCSE) support provides funding to the ARCHER user community to develop software in a sustainable manner to run on ARCHER.
There are currently no calls open. The list of all funded eCSE projects can be found on the funded eCSE projects page.
Useful Links
- List of all eCSE projects funded
- Codes worked on during eCSE projects
- eCSE Final report template
- Acknowledgement of eCSE funding
The eCSE programme provides tangible software enhancements to the communities exploiting software on ARCHER. This in turn has led to significant scientific advancements and both economic and social benefits to society.
Reports from completed eCSE projects
Implementation of Spin-Orbit Couplings in Linear-Scaling Density Functional Theory
Spin-Orbit Coupling (SOC) is a relativistic quantum-mechanical effect resulting from the interaction between the spin and orbital angular momentum of a particle. It has crucial effects on the electronic bandstructure of many technologically-relevant materials. While the underlying physics of how to implement it in calculations is well established, this functionality was not available in any linear-scaling, high-accuracy Density Functional Theory (DFT) package. In this project we implemented Spin-Orbit Couplings in a linear-scaling DFT code, ONETEP. This will significantly extend the range of materials that the linear-scaling code can describe accurately, and will lead to an enlargement of its user base. Read more...
A fully Lagrangian dynamical core for the Met Office NERC Cloud Model
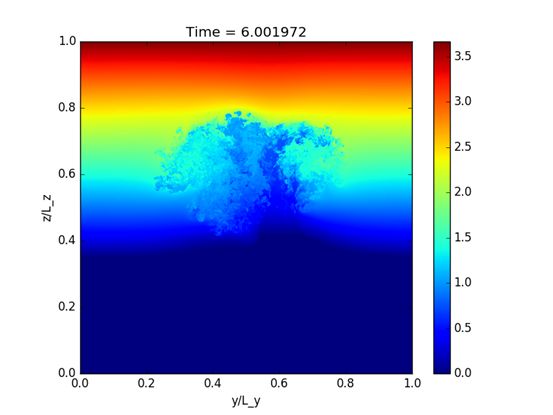
The turbulent behaviour of clouds is responsible for many of the uncertainties in weather and climate prediction. Weather and climate models fail to resolve the details of the interactions between clouds and their environment and suffer from a crude representation of microphysical processes, such as rain and snow formation. These processes can be studied in high-resolution "Large Eddy Simulations" (LES) (using a grid spacing below 100 metres) where the interaction between clouds and the environment is approximately resolved. Read more...
Developing Dynamic Load Balancing library for wsiFoam
Offshore and coastal engineering fields are using increasingly larger and more complex numerical simulations to model wave-structure interaction (WSI) problems, in order to improve understanding of safety and cost implications. Therefore, an efficient multi-region WSI toolbox, wsiFoam, is being developed within an open-source community-serving numerical wave tank (NWT) facility based on the computational fluid dynamics (CFD) code OpenFOAM®, as part of the Collaborative Computational Project in Wave Structure Interaction (CCPWSI). Read more...
Optimising CASTEP on Intel's Knight's Landing Platform
CASTEP is a widely-used, UK-developed software package, capable of predicting the properties of materials from "first-principles"; that is, by solving quantum mechanical equations to determine what the behaviour is, without the need for adjustable parameters. CASTEP was designed from the beginning to run well on conventional parallel HPC machines, but in recent years a number of new computer architectures have emerged which do not follow the conventional trends for CPUs. One such architecture is Intel's Knights Landing (KNL). Read more...
Efficient adsorption studies with DL_MONTE
Grand-canonical Monte Carlo (GCMC) is a widely-used simulation method for studying adsorption. DL_MONTE [1,2] is a general-purpose Monte Carlo program which can be used to study adsorption using GCMC in a wide range of systems, including those of relevance to the fields given above. DL_MONTE is part of the suite of simulation software developed by Daresbury Laboratory and the Collaborative Computational Project 5 (CCP5), and is accompanied by a Python toolkit for managing simulation workflows and performing data analysis. Read more...
PANDORA Upgrade: Particle Dispersion in Bigger Turbulent Boxes
PANDORA is a pseudo-spectral flow solver providing solutions to the Navier-Stokes equations in a cubic domain for isotropic and homogenous turbulent flow. It also includes the tracking of a large number of point, Lagrangian particles. A new version of PANDORA has been implemented within this eCSE project. The main aim of the project was to overcome the limitations of the previous code in terms of memory use and efficiency. Read more...
Full parallelisation and modernisation of the BGS global magnetic field model inversion code
The BGS global geomagnetic model inversion code is used to produce various models of the Earth's magnetic field. It is essentially a mathematical model of the Earth's magnetic field in its average non-disturbed state. The input consists of millions of data points collected from satellite and ground observatories on or above the surface of the Earth which are used to identify the major sources of the magnetic field: the core, crust, ionosphere and magnetosphere. Read more...
Development of a non-equilibrium gas flow capability within Code_Saturne using the Method of Moments
In this project, the functionality of the open-source CFD software Code_Saturne has been extended to enable the research community to investigate and predict non-equilibrium gaseous transport in complicated geometries in real engineering applications. For example, flow past a cluster of randomly distributed cylinders is often used to study the permeability of porous media. Read more...
Hybrid parallelisation for the CRYSTAL code
This project implements hybrid parallel programming concepts to improve the performance of the ab initio quantum-mechanical software CRYSTAL on modern supercomputers. Read more...
Grids in grids: hierarchical grid generation and decomposition for a massively parallel blood flow simulator
Implementing an accurate, consistent, parallel, efficient meshing tool is a very difficult software engineering task. We have created a tool that is accurate and consistent after much effort. Some key parts of the process are parallel and efficient and, for realistic problems, we can now produce voxelisations a factor of ten faster using a 36-core machine. Read more...
A fast coupling interface for a high-order acoustic perturbation equations solver with finite volume CFD codes to predict complex jet noise
In this project, our aim was to develop a fast interface for a high-order acoustic perturbation equations solver, APESolver, to couple with finite volume Navier-Stokes equations codes that are widely used in the UK Turbulence, Applied Aerodynamics and ARCHER communities. Driven by more stringent design requirements, the demand for multi-physics modelling capabilities is growing fast. Well-established modelling tools for individual disciplines are in need of running simultaneously while exchanging coupling data efficiently. Read more...
An adjoint solver for variable-density flows in the low Mach number limit
Incompact3D is a high-order, highly scalable code for simulating constant density flows. The high scalability is vital to be able to effectively utilise modern computing environments such as ARCHER to study complex flows, with applications ranging from fundamental turbulence research to investigating drag-reduction devices and wind farms. Incompact3D is based on an assumption of constant density, which limits its applicability to various flows of interest that may feature significant density variations, for example environmental flows such as dispersal of pollutants or avalanches. Read more...
Implementation of multi-level contact detection in granular LAMMPS to enable efficient polydisperse DEM simulations
Granular materials occur in many physical and industrial settings, such as geomaterials, avalanches and landslides, crushing of mining ores, food processing and pharmaceuticals. In many cases the grains/granules cover a wide range of particle sizes. For example, the granular filter material used to construct the Bennett Dam in Canada contains particles ranging from 0.08 to 75 mm.
Discrete element modelling (DEM) is a computational tool for predicting how such granular materials will respond during loading, flowing or other processes found in nature or in industry.
Read more...
Project Meshes - a HPC geometry library supporting mesh-to-mesh projections
Many mesh-based high performance applications require users to map their data from one discretised geometric representation (mesh) to another mesh multiple times.
The Delta library is a tool to handle imports/exports of geometries
for use cases as the one above. Realising the above features is usually time consuming and technical. Our code serves as building block to simplify setting up realistic simulations using realistic geometries.
Read more...
Massively Parallel MPI Implementation of the SPH Code DualSPHysics
Smoothed Particle Hydrodynamics (SPH) is a novel computational technique that is fast becoming a popular methodology to tackle industrial problems with violent flows that often occur in nature and industry. The main aim of this project is the implementation of MPI functionality to the open-source DualSPHysics software package, developed by the universities of Manchester, Vigo and Parma, enabling the simulation of violent, highly transient flows using thousands of cores such as slam forces due to waves on offshore and coastal structures, impact loads on high speed ships, ditching of aircraft, sloshing of fuel tanks and many more. Read more...
Implementation of generic solving capabilities in ParaFEM
In this eCSE project, the PETSc solver library has been interfaced to the ParaFEM finite element analysis (FEA) library.
FEA is widely used in engineering to simulate the deformation of structures under load. In FEA, the structure is discretised into small elements (often millions of these), each connected to its neighbours. This leads to a large system of equations to solve and at the heart of every FEA program is an equation solver. The orthopaedic engineering research group at the University of Edinburgh uses the ParaFEM FEA library to simulate the mechanical behaviour of trabecular bone, using a detailed geometry generated through X-ray microtomography in order to assess the response of bone to mechanical loads at the microstructural level Read more...
Discrete velocity methods for helicopter multi-block CFD solver: towards real-time wake simulations
This project is a step towards the development of real-time simulations using detailed flow physics. Such simulations are necessary to enhance the physics included in the models used for flight simulators. An example is the simulation of a helicopter operation near a building, ship, or wind turbine where due to wind, the resulting wakes are affecting the performance and the operation of the helicopter. Read more...
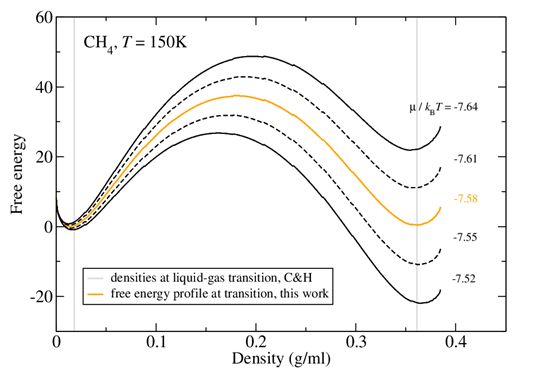
Implementing lattice-switch Monte Carlo in DL-MONTE to unlock efficient free energy calculations
The Monte Carlo technique - so called in a nod to the Monte Carlo Casino in Monaco - makes use of chance or probability to investigate the properties of many important materials: metals, minerals, and compounds both fluid and solid. The chance is introduced by selecting random rearrangements of the atoms or molecules and then computing how likely such a rearrangement might be in a real system. As there is significant freedom in how to choose the random rearrangement, a number of different Monte Carlo techniques exist. Read more...
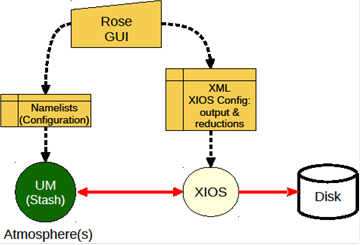
Implementation of XIOS in the Atmospheric Component of the Unified Model
In the Met Office Unified Model, used extensively in the ac.uk community, the diagnostic system, while very rich in capability, is tightly integrated with the model dynamics and physics. Increasing use of couplers (as for example between NEMO (an ocean model) and the atmospheric UM) leads naturally to the idea that the diagnostic system itself could (should) be a coupled component of a climate model.Our operational familiarity with XIOS and NEMO led us to propose XIOS as a candidate a coupled diagnostic component for the UM. Read more...
Preconditioned Geometry Optimisers for the CASTEP and ONETEP codes
Density functional theory (DFT) is one of the most widely used computational methods to investigate molecules and materials by performing calculations that explicitly the electronic degrees of freedom. The CASTEP and ONETEP DFT packages are UK flagship codes and are heavily used on ARCHER (making up 8 and 1% respectively of recent usage). The application of preconditioners requires solving a linear system, which is the most expensive part of their application. However, our aim was to keep the computational cost of the construction of the preconditioners and the solution of linear system below 5% of the cost of the electronic structure problem. Read more...
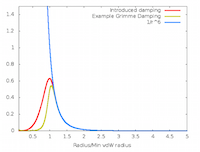
Enhancing long-range dispersion interaction functionality of CASTEP
In this project the semi-empirical dispersion correction (SEDC) schemes were refactored to improve their sustainability, enabling alternative methods to be implemented much more straightforwardly. By linking with a new software library, the newer D3 and D4 schemes of Grimme were added to CASTEP. The MBD was also refactored and re-implemented, improving its performance, accuracy and parallel scaling substantially (often by more than an order of magnitude). Read more...
Enabling NAME for distributed memory parallelism
The Numerical Atmospheric dispersion Modelling Environment (NAME) is owned by the United Kingdom Met Office and is used to perform simulations of a wide range of atmospheric dispersion phenomena. These include, but are not limited to: nuclear accidents, volcanic eruptions, chemical accidents, smoke from fires, and transport of airborne disease vectors.However, as the resolution of meteorological data increases, the number of particles required to maintain statistical accuracy also increases. The associated increase memory and computational requirement then favours a distributed memory implementation to complement the existing shared memory implementation. Read more...
CVODE support for FEniCS
FEniCS is a widely used platform for solving partial differential equations (PDEs). PDEs describe physical laws, such as how heat travels, or magnetisation varies in a magnetic specimen. In this project, we have extended FEniCS to support the SUNDIALS CVODE . SUNDIALS CVODE provides methods for solving systems of ordinary differential equations, which when coupled to FEniCS provides a wide range of methods for solving time-dependent partial differential equations. Read more...
TPLS: 3D Decomposition and Gas/Liquid Flows
TPLS is a freely available CFD code which solves the Navier-Stokes equations for an incompressible two-phase flow. This eCSE project aimed to improve the architecture and performance of the software, reduce overall runtime, and improve usability and maintainability, opening up the software to a large user community. Read more...
Extending CPL library to enable HPC simulations of multi-phase geomechanics problems
The project has enabled the simulation of fluid-particle systems by combining the molecular dynamics code LAMMPS and the computational fluid dynamics code OpenFOAM on ARCHER. The resultant system will be of use to engineers in geotechnical (civil) engineering, geology, and petroleum engineering. Read more...
CP2K - Electron Transport based on Non-Equilibrium-Green's-Functions Method
This project will add to CP2K a Density Functional Theory based Non-Equilibrium Green's Functions (NEGF) method for simulating quantum electron transport in nano-scale systems. It allows the use of full set of DFT functional and van der Waals dispersion interaction models currently available to CP2K. Read more...
Task-Farming Parallelisation of Py-ChemShell for Nanomaterials
This project aims to parallelise QM/MM calculations in the newly-developed version of ChemShell written in the Python programming language. During the project we have successfully completed the code development for performing "task-farmed" parallel calculations which allows us to model very large scale chemical systems. We have carried out benchmark calculations to demonstrate that the computational efficiency can be greatly improved using our implemented code. Read more...
Optimizing the I/O performance of OpenFOAM for massively parallel high-fidelity CFD simulations
OpenFOAM is a comprehensive, flexible, widely used package for CFD computation. Whilst OpenFOAM has many strengths it's code design has resulted in some fundamental weaknesses for I/O. The aim of this project was to prototype the introduction of a modern, parallel, large scale I/O into OpenFOAM. Read more...
Developing Massively Parallel ISPH with Complex Boundary Geometries
The incompressible Smoothed Particle Hydrodynamics (ISPH) method with projection based pressure correction has become very attractive due to its high accuracy and stability for both internal flow and free surface flows. The main aim of this project is to develop new functionalities which are used to enable to simulate multiple floating bodies with a very large number of particles using tens of thousands of cores. Read more...
Large scale CASTEP calculations to interpret solid-state NMR and Vibrational Spectroscopy Experiments
In this project we have significantly improved the parallel scaling of the spectroscopic modules in CASTEP (specifically NMR and vibrational) by introducing an additional layer parallelism into the code - parallelism over perturbations. Many spectroscopic parameters can be considered as the response of the electronic structure to a perturbation. For example the NMR J-coupling is the response of a system to the magnetic field generated by excitation of a nuclear spin; the NMR magnetic shielding is the response of a system to an applied external magnetic field; lattice dynamics is the response of the system to the movement of the ionic cores. Read more...
Implementation and optimisation of advanced solvent modelling functionality in CASTEP and ONETEP
Implicit solvent models provide a simple, yet accurate means to incorporate solvent effects into electronic structure calculations. In this project we have implemented one such model, the minimal parameter solvation model (MPSM), in two electronic structure packages: CASTEP and ONETEP. In the MPSM, the electrostatic potential which describes the interaction of the implicit solvent and solute is determined by direct solution of the nonhomogeneous Poisson equation (NPE). Read more...
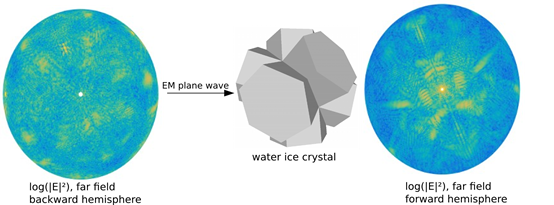
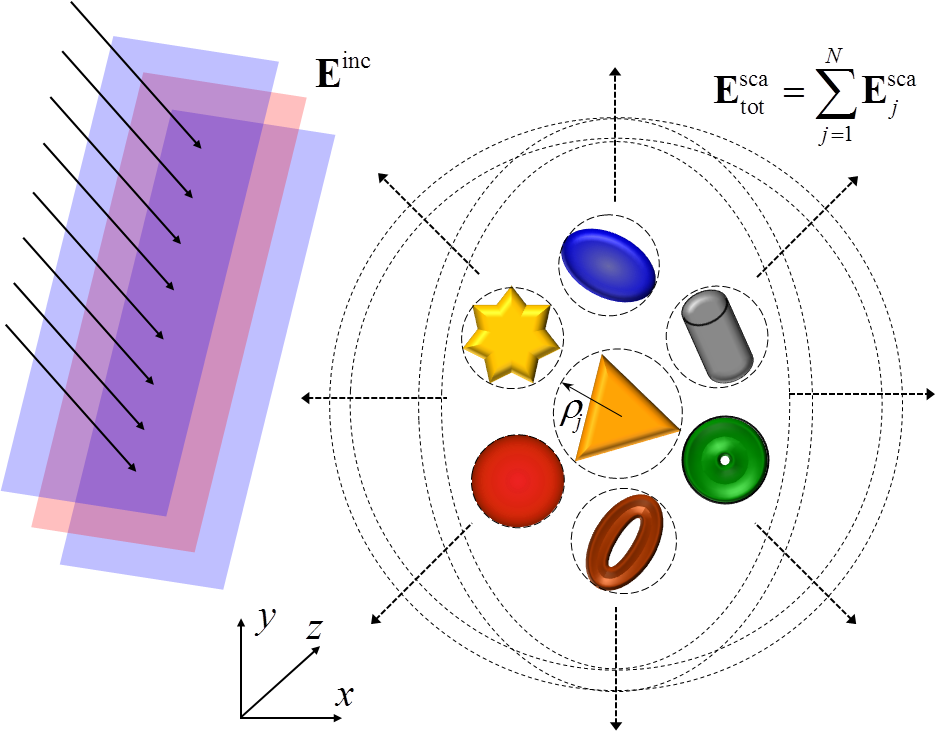
ADDA: Efficiency of computations of electromagnetic scattering from arbitrary particles
The main objective of the project was to make computations of electromagnetic scattering properties of small particles and structures of arbitrary shape more accessible to future users, and to ensure that the resources are used efficiently. This involved implementation, modernization and improvements to the publicly available open source version of the Discreet Dipole Approximation technique ADDA, providing user assistance through documentation, examples and a user interface to set up runs and providing post processing tools needed in atmospheric application. Read more...
Distributed Hamiltonian build and diagonalisation in UKRMol+
The goal of the project was to completely rewrite the inner region Hamiltonian build and diagonalizer code SCATCI to enable its usage on distributed computing clusters. The rewrite was chosen as opposed to simply a port as the Fortran 77 used limits its readability and expandability for future work. Modern Fortran (2003+) was chosen to allow current users to continue maintaining the code whilst leveraging the powerful features of object-oriented programming (OOP). Read more...
VAMPIRE: Billion-atom simulations of magnetic materials
The aims of this eCSE project were to optimise the VAMPIRE code on the ARCHER system and improve the data input/output routines to enable configuration data to be extracted from the simulation to see the time evolution of the atomic spins. The improved code enables a new class of magnetic materials simulation containing between 10,000,000 and 1,000,000,000 atoms. Such large simulations give unprecedented insight into to the behaviour of complex magnetic materials for realistic situations. Read more...
Optimal parallelisation in CASTEP
When running a parallel CASTEP calculation using many threads, some tasks were performed on only a single thread. While most of this work takes a small portion of the runtime, as larger systems are studied the time taken by some of these routines becomes significant. This project aimed to distribute the work in these significant tasks across multiple threads, therefore significantly reducing the time taken.Read more...
Optimisation of LESsCOAL for large-scale high-fidelity simulation of coal
This project aims to substantially improve the parallel performance of an in-house code, LESsCOAL (Large-Eddy Simulations of COAL combustion). This has been under active development over recent years by one of the PI's main collaborators, Prof Zhihua Wang, working at the State Key Laboratory of Clean Energy Utilization of Zhejiang University, China. While originally designed to properly predict turbulent pulverised-coal combustion using high-fidelity simulation techniques, the software package has now developed to a stage at which it can be used for a variety of turbulent multiphase reacting flows.
Read more...Enabling large-scale microphysics in the MONC weather model
This project focused on a number of computational areas, including optimising the iterative solver which is used to solve the pressure equation. The traditional method used here inherently limits the scalability of the code. By integrating with a common and widely available solver toolkit, the user has a significant choice about which solver to use and how. We have also optimised the CASIM micro-physics scheme, used to model moisture interactions. Our work has more than doubled the performance of this code on CPUs. As part of this work, CASIM has also been ported to GPUs and KNL, to see if these novel architectures could help to optimise performance.
Read more...Integrating mesh movement technology within Fluidity and the PRAgMaTIc adaptive mesh toolkit
This work builds upon previous mesh adaptivity technology developed at Imperial College London which has primary focused on 'h' adaptive, or mesh optimisation, techniques. In this approach the shape/size of the elements that constitute an unstructured mesh are periodically updated in response to an evolving numerical simulation. The aim is to only use higher resolutions when and where they are needed, in order to optimise the accuracy and efficiency of a calculation. Read more...
Local Excitement in CP2K
The work in this eCSE project enables much more realistic models of systems a method, called Time Dependent Density Function Theory, within the open source atomistic simulation code CP2K. The reduced scaling (linear in system size) and smaller prefactor than previous implementations will allow excited state calculation to be performed more routinely, for larger systems, in a time-resolved manner along molecular dynamics trajectories and will be suitable for high-throughput applications. Read more...
Improving the ease-of-use and portability of the COSA 3D solver for open rotor unsteady aerodynamics
In this project, we aimed to significantly improve both the performance and usability of COSA, a Navier-Stokes (NS) Computational Fluid Dynamics (CFD)code designed for internal and external flow applications. We have made a number of improvements to COSA to improve the performance and usability of the code. A large part of our optimisation work focused on improving the I/O performance of COSA at scale, and we have reduced the cost of I/O at large core counts by as much as 70%, significantly reducing parallel overheads and saving simulation time. Read more...
An Exascale Multi-Scale Framework for Solid Mechanics
This project has delivered a scalable tool that can satisfy this need, initially for fracture problems. A multi-scale engineering modelling framework for deformation and fracture in heterogeneous materials (polycrystals, porous graphite, concrete, metal matrix or carbon fibre composites) has been developed. This has good potential for use on HPC systems, such Tier-1 ARCHER, Tier-2 Hartree centre systems, and smaller University and departmental systems. Read more...
Fast and Massively Distributed Electromagnetic Solver for Advanced HPC Studies of 3D Photonic Nanostructures
The work performed in this project built upon our recent development of the OPTIMET code, a project funded by the University dCSE Programme. OPTIMET is based on the multiple scattering matrix (MSM) method, a method widely used in computational electromagnetism. The main computational work in the MSM method consists of computing the system scattering matrix, S and solving a linear system of equations, Sa=b. Once a is known, the electromagnetic field can be computed at any point, as well as the corresponding cross-sections. As part of the eCSE project, the efficiency and functionality of the code were greatly enhanced. Read more...
libcfd2lcs: A general purpose library for computation of Lagrangian coherent structures during CFD simulation<
In this eCSE project, a new computational library, libcfd2lcs, was developed, that provides the ability to extract LCS on-the-fly during a computational fluid dynamics (CFD) simulation with only modest additional overhead. Users of the library create "LCS diagnostics" through a simple but flexible API, and the library updates the diagnostics in the background as the user's CFD simulation evolves. By harnessing the large-scale parallelism of platforms like ARCHER, this enables CFD practitioners to make LCS a standard analysis tool, even for large scale three-dimensional simulations. Moreover, libcfd2lcs provides access to the time evolution of LCS with unprecedented detail, giving researchers a powerful new tool to study unsteady flow structures. Read more...
Adding Multiscale Models of DNA to the LAMMPS Molecular Dynamics code
DNA modelling has been an important field in biophysics for decades. In this project, the oxDNA coarse-grained model for DNA and RNA was ported into the popular and powerful LAMMPS molecular dynamics code. This makes oxDNA widely accessible to the global community of LAMMPS users, whereas previously it was only available as bespoke and standalone software. Through the efficient parallelisation of LAMMPS it is now also possible to run oxDNA in parallel on multi-core, multi-processor and distributed memory architectures, extending its capabilities to unprecedented time and length scales. Read more...
Introducing Thread and Instruction Level Parallelism into Ludwig Code for Modelling Complex Fluids
The Ludwig code is used to simulate a wide variety of soft matter substances, including liquid crystals. Liquid crystals are widespread in technology (including displays and other optical devices), but much is yet to be understood about the range of possible configurations. Simulations are vital in paving the way to improved knowledge. This project applied a new custom-made programming model, targetDP, to Ludwig to allow it to perform as well as possible not only on ARCHER but across a range of emerging hardware platforms, thereby exploiting the more complex hierarchical parallelism which is becoming typical. Read more...
Large-Eddy Simulation Code for Air Quality Modelling in City-Scale Environments
The ELMM large-eddy simulation software is used for simulating wind flow in the atmospheric boundary layer, and the transport and dispersion of pollutants in areas with complex geometry, primarily cities. This project aimed to enhance the code to enable simulations of whole cities, i.e. areas with a horizontal dimension of 10 km or more, while maintaining high resolution in the areas of interest. The work has enabled much larger simulations to be carried out in areas of great interest such as urban air quality, including modelling of the dispersion of dangerous pollutants in cities and of the effects of the urban heat island. Read more...
Developing Fluidity for high-fidelity coastal modelling
Fluidity is an open source finite-element computational fluid dynamics code. This project aimed to improve the performance of Fluidity and improve its ease of use. The improvements made to the code allow fluids problems to be studied in greater detail, and open up Fluidity to tidal modellers, with particular applications in the field of marine renewable energy. Research in areas such as aerodynamics, wind energy, marine energy, and environmental/pollution modelling will also benefit from the improvements to the code. Read more...
Implementation of a highly scalable aeroacoustic module in Code_Saturne Computational Fluid Dynamics software
This project aimed to expand the capabilities of the open-source CFD software Code_Saturne to directly perform noise predictions by means of an acoustic analogy. Noise prediction is key for many engineering applications ranging from aerospace to combustion, but traditional methods of simulating noise are very computationally demanding. This project aimed to implement an acoustic module within Code_Saturne while maintaining the code's excellent scalability. Read more...
Removing pseudo-linear dependence in Gaussian basis set calculations on crystalline systems with CRYSTAL
CRYSTAL is a world-leading electronic structure program for the ab initio quantum mechanical simulation of crystals, nano-structures, surfaces and molecules. Its uses include research into new materials in areas such as photovoltaics. This project implemented automatic screening of the matrices affected by linear dependence issues when using high-quality molecular basis sets in CRYSTAL. By reducing the effort associated with the calculation set-up and with convergence checks for large basis sets, the results enable a wider community of users to perform faster calculations and achieve higher levels of accuracy in the calculated properties. Read more...
Implementation of Dual Resolution Simulation Methodology in LAMMPS, leading to improved studies of biological processes
Classical Molecular Dynamics simulations are widely used to understand the behaviour of biological systems. Calculations at the atomistic level are very computationally demanding, but coarse-grain (CG) models provide less accuracy. Hybrid models allow the most critical parts of the system to be represented at the atomistic level, with the remainder done by CG. This project updated and optimised the LAMMPS simulation software, enabling the use of hybrid simulations for the study of larger systems over longer timescales. This will not only improve our understanding of important fundamental biological processes, but also has the potential to assist with the development of new drug therapies. Read more...
Full parallelism of optimal flow control calculations with industrial applications
This project aimed to fully parallelize SEMTEX, a well-established open-source quadrilateral spectral element Direct Numerical Simulation (DNS) code which previously only had a limited parallel implementation. The improvements to the code will accelerate research in developing disruptive technologies in flow control, which is recognised as the leading edge of fluid dynamics research. The code has applications in the oil, aviation and wind energy industries. Read more...
Enabling high performance computing for tools for the analysis of single molecule ion channel currents: probing the energy landscape of channel activation to understand the protein's structure-function relation
Ion channels mediate excitability and information processing in all living organisms and are the target of many therapeutic drugs. Recent technical advances have enabled the recording of the tiny (picoampere) electric current flowing through a single ion channel. Uniquely for a protein, tens of thousands of channel openings and closings with a temporal resolution of around 10 microseconds can be recorded in a single experiment. HJCFIT is a library for the maximum likelihood fitting of kinetic mechanisms to these sequences of open and shut time intervals. This project has enabled HJCFIT for ARCHER, significantly shortening the analysis time, potentially allowing more complex questions to be addressed and enabling a new community of researchers who have not previously exploited HPC facilites for the efficient use of HJCFIT. Read more...
POpSiCLE: A photoelectron spectrum library for laser-matter interactions
Molecules driven out of equilibrium by intense, ultrashort laser pulses are of central importance in many areas of science and technology. In such a non-equilibrium situation, charge and energy transfer can be induced across the molecule on a femtosecond timescale. Understanding and controlling these transfer processes is fundamental to many chemical processes and key to future ultrafast technologies: e.g. the design of electronic devices, probes and sensors, biological repair and signalling processes, and development of optically driven ultrafast electronics. This project developed a portable parallel library that implements three methods for calculating photoelectron spectra. Read more...
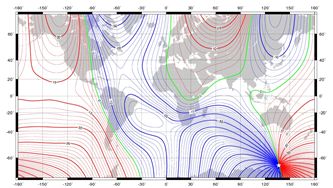
Improving the EMPIRE data assimilation code for weather forecasting and other geosciences applications
Data assimilation is an essential part of any prediction system, such as weather forecasting. It combines a numerical model with all observations available to generate the best starting point for the forecast. The EMPIRE data assimilation system is a community system used by increasing numbers of researchers across the UK from all branches of geosciences. This project improved the efficiency of the EMPIRE software. The improved code will be available to all EMPIRE users on ARCHER and future UK national HPC systems. Read more...
Enabling high-performance oceanographic and cryospheric computation on ARCHER via new adjoint modelling tools
The study of the ocean and cryosphere is crucially dependent on computationally-intensive, large-scale numerical models that must be run on HPC systems. In recent years, the use of adjoint models has led to unique insights in this area. However, the uptake of adjoint modelling has been hindered due to poor access to efficient, cost-effective tools. This project aimed to bolster the ARCHER community's access to oceanic and cryospheric adjoint modelling through the development of model test suites. Read more...
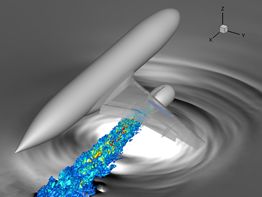
Optimising Essential Particle Dynamics Codes for Multi-Scale Engineering Flow Simulation
This project aimed to further develop a software application built upon the Computational Fluid Dynamics (CFD) framework OpenFOAM, which uses the framework to run molecular dynamics (MD) simulations of nanoscale and multi-scale flow problems. The output software of this project has had a substantial impact in generating scientific insight for future micro/nano flow technologies with many practical applications, e.g. water impedance in carbon nanotube membranes for water purification; slip flows over nanostructured surfaces for marine drag-reduction, and control of deposit patterns using electro-wetting of nano-droplets for 3D printing. Read more...
Image courtesy of Micro & Nano Flows for Engineering
QuasiParticle Self-Consistent GW electronic structure calculations of many-atom systems
Quasiparticle Self-Consistent GW approximation (QSGW) is a novel method for electronic structure calculations, which addresses many shortcomings of the standard method, density functional theory (DFT). An example of its use is for studies of photovoltaic materials and their efficiencies. QSGW is applied successfully to small systems. This project aimed to enable QSGW calculations for larger system sizes, by parallelising the code and adapting it to large multi-core HPC systems. Read more...
Improving DL_POLY_4 molecular dynamics package performance and scalability using multiple time stepping
DL_POLY is a general purpose classical molecular dynamics (MD) simulation software package, which is used by the materials science, solid state chemistry, biological simulation and soft condensed-matter communities. In this project, researchers at STFC and the University of Oxford have implemented a multiple time stepping scheme, RESPA, for less frequent calculations of expensive operations. This has been shown to consistently bring about a 15-25% improvement in performance at fixed process count, and noticeably improves the scaling of the code. Read more...
High performance multi-physics simulations with FEniCS/DOLFIN
The FEniCS Project is a collection of free, open-source, software compnents with the common goal of enabling automated solution of differential equations. DOLFIN is a C++/Python library which functions as the main problem-solving environment and user interface. Researchers at the University of Cambridge have enhanced the FEniCS/DOLFIN library to extend its functionality for multi-physics / multi-field simulations on parallel computers. The new functionality has enabled simulations of partially molten magma and analysis of CO2 subsurface sequestration to be carried out. Future projects which will benefit from the the improvements include brain simulations and simulation of float glass processing. Read more...
Delivering a step-change in the performance and functionality of the Fluidity shallow water solver
Fluidity is an open source, computational fluid dynamics (CFD) framework used to solve large-scale geophysical/oceanographic problems, such as marine renewable energy, tsunami simulation and inundation, and palaeo-tidal simulations for hydrocarbon exploration. Researchers from Imperial College London have significantly enhanced Fluidity's shallow water model, and prepared the code for running on state-of-the-art HPC systems without the need for model developers to re-write any of their existing code. Preliminary work on tidal turbine modelling was performed during this project and the improved code has also been used in an undergraduate project to study the potential impact of tsunamis to the UK. Read more...
Parallel supermeshing for multimesh modelling
Models which use multiple non-matching unstructured meshes typically need to solve a computational geometry problem, and construct intersection meshes in a process known as supermeshing. An implementation of the algorithm for solving this problem exists in the unstructured finite element model Fluidity, but this implementation has, until now, been deeply embedded within the code and unavailable for widespread use. In this project, researchers from the University of Edinburgh and University of Oxford created a standalone general-purpose numerical library, named libsupermesh, which can easily be integrated into new and existing numerical models. Read more...
VOX-FE for biomechanical modelling - new functionality for new communities
Understanding the mechanics of biological structures such as bones is very challenging, and experimental measurements can be extremely difficult. Computational modelling offers an alternative approach for studying the biomechanics of living and extinct animals, from insects to dinosaurs. In this project, researchers at the Universities of Hull and Edinburgh have significantly extended the VOX-FE modelling software, which can be used in predictive biology and virtual experimentation to reduce animal experiments. Potential applications include high resolution modelling of whole bones to understand normal and pathological bone biomechanics (e.g. in osteoporosis), and in silico design and testing of new dental and orthopaedic implants. Read more...
Optimisation of the EPOCH laser-plasma simulation code
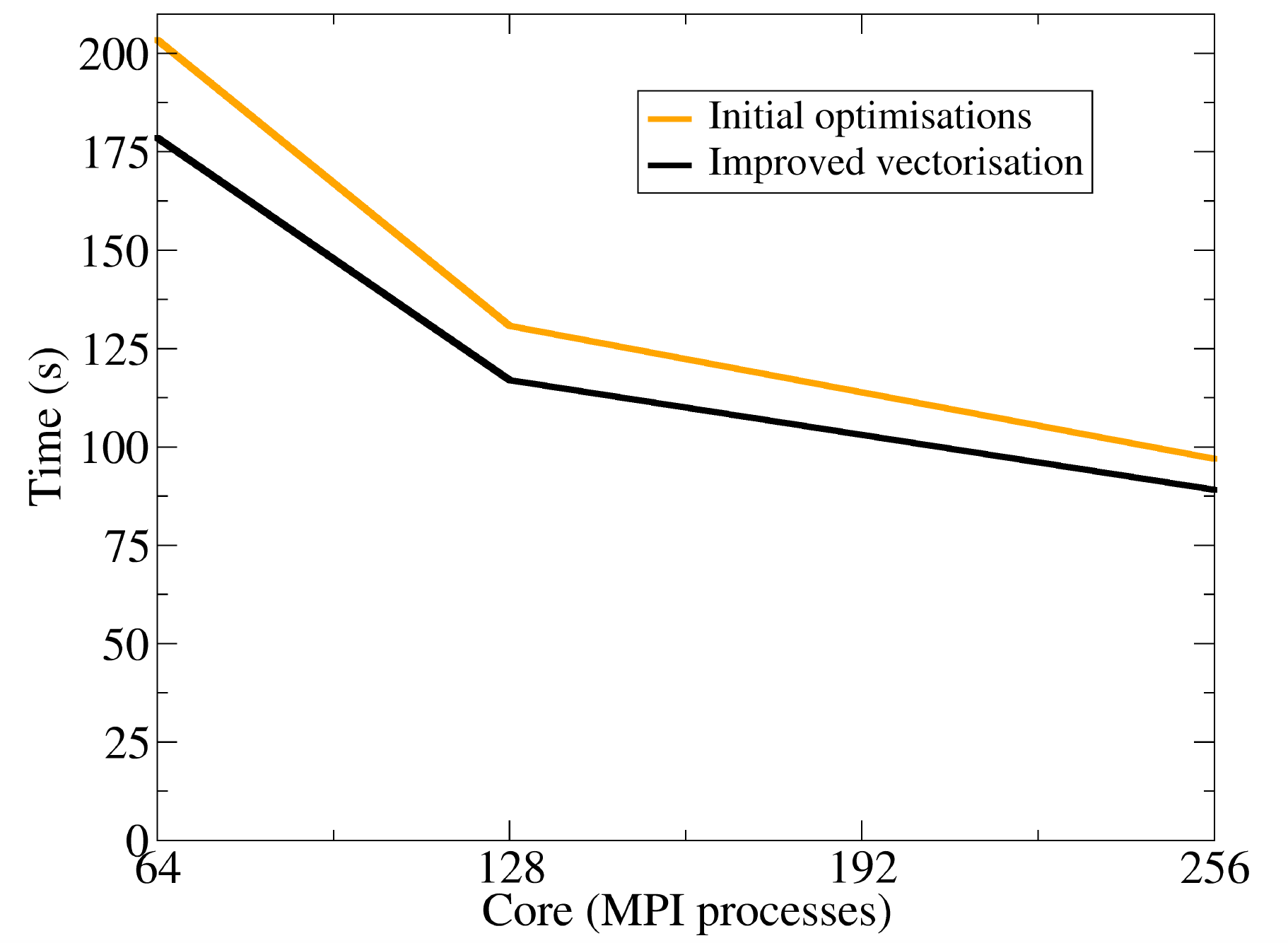
The EPOCH laser-plasma simulation code is a mature community code with a large international user base. The code uses particle-in-cell (PIC) techniques. The purpose of this project was to both improve the performance of EPOCH and futureproof the code for next generation multi-core architectures. As a result of the improvements made to the code, a speed-up of up to 3.58 has been achieved, and EPOCH can now be used for a wider variety of simulations that feature non-uniform particle distributions. Read more...
Image: Illustration of Hilbert space-filling curves for 3D spaces: William Gilbert, University of Waterloo
NEMO Regional Configuration Toolbox for ocean modelling
NEMO is a state-of-the-art ocean modelling framework used by the Met Office, NERC and many other UKHEIs to simulate the ocean from the global scale down to that of estuaries. This project has developed a unique set of tools - the NEMO Regional Configuration Toolbox (NRCT) - to aid users to set up lateral boundary conditions for near real-time regional ocean simulations within the NEMO environment. The NRCT provides access to remote datasets, and the tool effectively translates data from a parent model source into a format suitable to run a regional NEMO ocean simulation. A GUI is also provided to allow the user to define the regional ocean model domain. Read more...
Improving the performance of Nektar++ using communication and I/O masking
Nektar++ is an open-source spectral/hp element framework for numerical solution of partial differential equations in complex geometries using high-order discretisations. As a result of the work done in this project, the code now makes more efficient use of computational resources and scales to higher core counts, allowing simulations to run in a shorter period of time. This benefits a wide range of application areas including transient aerodynamics simulation (F1, aircraft design and analysis), aero-acoustic simulation (aircraft engines), modelling of patient-specific cardiac electrophysiology (atrial fibrillation) and biological flow simulations for understanding disease formation and development (atherosclerosis). Read more...
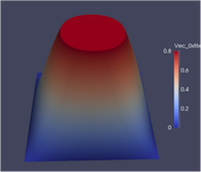
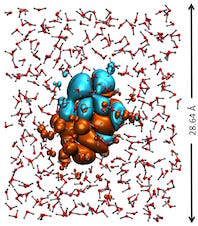
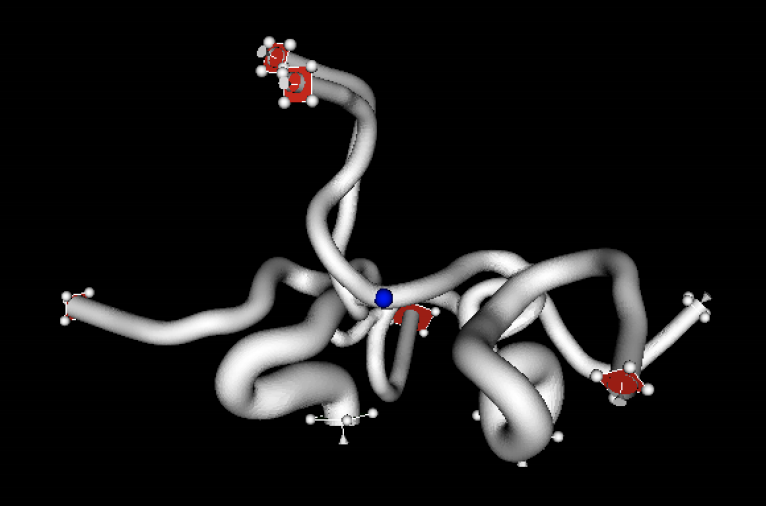
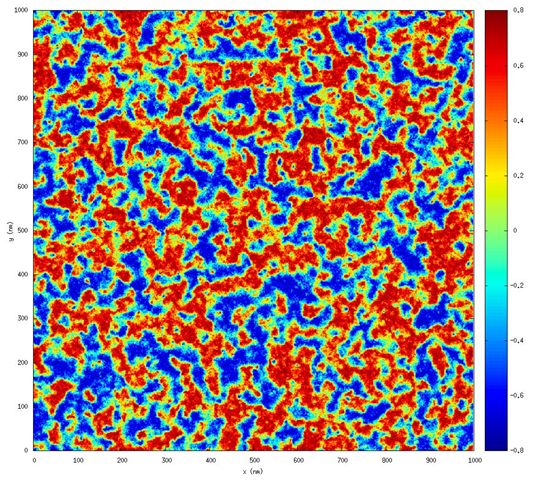
Algorithmic Enhancements to the Solvaware Package for the Analysis of Hydration
The Solvaware package is a workflow that runs and analyses molecular dynamics (MD) trajectories to estimate hydration free energies by computing the contribution of different subvolumes around a solute. The accurate calculation of hydration free energies is a vital goal in computational modelling of biological and engineered aqueous systems. This project has enhanced the Solvaware package by improving the efficiency of key algorithms in the workflow, increasing its functionality, and widening its impact by making it available to the ARCHER community. Read more...
Optimising GS2 plasma physics simulation code used by fusion energy researchers
This project improved the performance at high core counts of the GS2 plasma physics simulation code. With a speed-up of up to 5 times (for short simulations) or up to 2 times (for longer simulations), this will allow plasma simulations to be carried out in much more detail, leading to a better understanding of the turbulence features in the plasma. Understanding, and controlling, turbulence in plasma is one of the key requirements for building a usable fusion reactor, and codes like GS2 can help in the design and understanding of such reactors. Fusion energy has the potential to provide a clean source of electricity for future generations. Read more...
Developing highly scalable 3-D Incompressible Smoothed Particle Hydrodynamics
Smoothed Particle Hydrodynamics (SPH) is a novel computational technique that is fast becoming a popular methodology to tackle industrial problems with violent flows. Recently, accurate incompressible SPH (ISPH) codes have become more popular now that the numerical stability can be ensured, particularly for free-surface flows. In this eCSE project, researchers from the University of Manchester and STFC Daresbury Laboratory have developed a scalable, 3-D ISPH engineering toolkit. Read more...
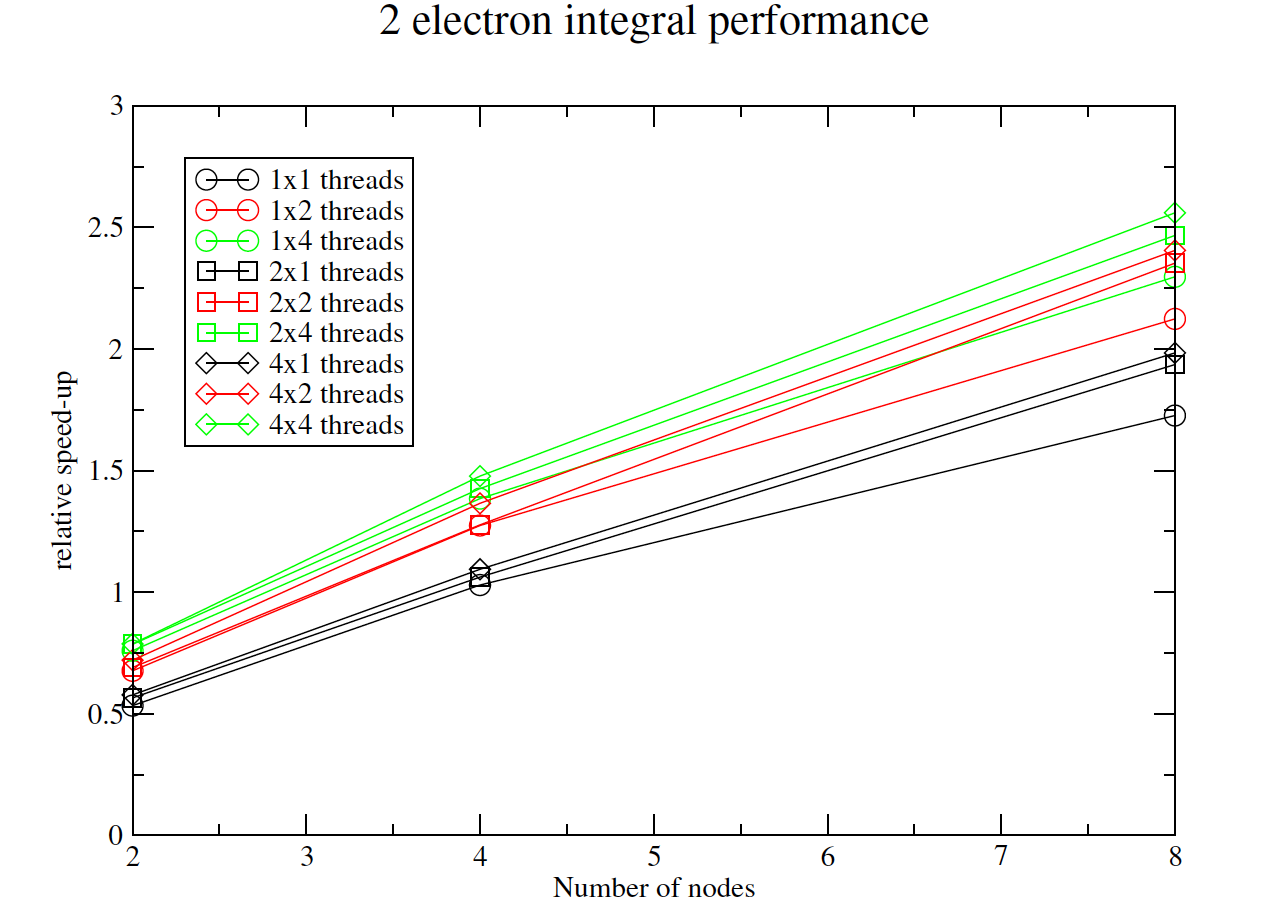
Efficient computation of two-electron integrals in a mixed Gaussian/B-spline basis
The aim of this project was to develop efficient parallel routines to calculate two-particle integrals in a mixed basis of Gaussian Type Orbitals (GTOs) and B-spline orbitals. The object-oriented library developed in the project has contributed to a significant improvement in the capability of the UKRmol+ suite of codes, designed to study electron and positron collisions with molecules. This will allow the study of electron interaction with bigger molecules than ever before, which could lead, for example, to a better understanding of how radiation damages cell constituents, in particular DNA. Read more...
Image: DNA ©iStock.com/nechaev-kon
Optimising van der Waals simulations with CASTEP code
CASTEP is a general purpose code for the quantum simulation of properties of materials. Researchers have improved the performance of the van der Waals (vdW) interactions as implemented by CASTEP by replacing the previous simple sum method with a modified Ewald scheme. The code has also been parallelised using MPI. As a result of these two changes, for large calculations on ARCHER the code now runs many times faster. Read more...
Microvasculature blood flow simulation using HemeLB
The work done in this project by researchers at UCL represents a substantial leap forward for the simulation of blood flow in microvasculature and enables for the first time the theoretical study of advanced aspects of haemorheology, oxygen transport, and cell trafficking in realistic vascular networks, and of the collective dynamics of dense red blood cell suspensions in complex vessel geometries. Read more...
Pushing the Frontiers of Kinetic Monte Carlo Simulation in Catalysis - Zacros Software Package Development
The importance of heterogeneous catalysis in applications that enhance the quality of life cannot be overstated. Iron-based catalysts for the production of ammonia by the Haber process is a notable example, the importance of which is shown by the fact that fertilisers generated from ammonia produced in this way are responsible for sustaining one third of the Earth's population. Scientists at UCL have employed kinetic Monte Carlo methodology and their own software package, Zacros, to contribute to the unravelling of complex phenomena on catalytic surfaces. This knowledge can be used to improve existing catalysts or design novel materials with optimal activity and selectivity for bespoke applications. Read more...
Scalable automated parallel PDE-constrained optimisation for dolfin-adjoint
The dolfin-adjoint package enables the automated optimisation of problems constrained by partial differential equations (PDEs). These problems are ubiquitous in engineering, eg the design of wings to maximise lift, or the cheapest bridge that will support the required load. Prior to this project, dolfin-adjoint was limited to serial optimisation libraries with no concept of parallel linear algebra. The work done in this project has eliminated the performance penalty associated with gathering the data and lifted the upper bound on the size of problems which can be considered. Read more...
Hybrid OpenMP and MPI within the CASTEP code
First-principles simulations of materials have had a profound and pervasive impact on science and technology, from physics, chemistry and materials science to diverse areas such as electronics, geology and medicine. CASTEP is a widely-used, UK-developed program for the quantum mechanical modelling of materials. This project has added a new level of parallelism to CASTEP, laying the foundations for a version of the code that will work well on future exascale supercomputers. The net result is a new science capability, allowing the study of larger and more complex systems than before, in less time. Read more...
Tuning FHI-Aims for complex simulations on Cray HPC platforms
The main objective of this project was to tune FHI-aims, a quantum mechanical electronic structure code, for optimal efficiency on ARCHER's hardware, for the benefit of members of the Materials Chemistry Consortium (MCC) and other users. The overarching target of this work by scientists at University College London was to improve scalability for a wide range of applications to enable new science, using current and emerging UK computer facilities, in a wide range of fields including materials and life sciences, chemistry, physics, and engineering. Read more...
Calculating Excited States of Extended Systems in LR-TDDFT
Electronic structure theory is a hugely important contributor to understanding properties of materials. Large-scale Density Functional Theory (DFT) codes such as ONETEP allow us to predict ground-state properties for large systems such as complex biomolecules. In this project, scientists at the University of Cambridge aimed to enhance, extend and improve the implementation in ONETEP of Linear Response Time-Dependent Density-Functional Theory (LR-TDDFT), the method of choice for computing optical properties of large systems. Read more...
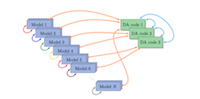
Porting and enabling use of the Community Earth System Model (CESM) climate model on ARCHER
The Community Earth System Model (CESM) is a state-of-the-art coupled climate model for simulating the earth's climate system. Composed of four separate sub-models simultaneously simulating the earth's atmosphere, ocean, land surface and sea-ice, and one central coupler component, CESM allows researchers to conduct fundamental research into the earth's past, present and future climate states. Scientists at the University of Edinburgh have ported to ARCHER and optimised two versions of CESM, and these are now both readily usable by the UK climate research community. This is expected to generate a further growth in interest in the model. Read more...
A pinch of salt in ONETEP's solvent model
Chemical reactions, drug-protein interactions, and many chemical and physical processes on surfaces are examples of technologically important processes that happen in the presence of solvents. The inclusion of electrolytes (salt) in solvents such as water is crucial for biomolecular simulations, as most processes (e.g. protein-protein or protein-drug interactions or DNA mutations) take place in saline solutions. This project aimed to develop the capability to model electrolyte-containing solvents in quantum-mechanical simulations of materials from first principles. Using a linear-scaling code such as ONETEP enables simulations to be performed on entire biomolecules or catalysts that typically involve hundreds or thousands of atoms. Read more...
Performance enhancement in RMT codes in preparation for application to circular polarised light fields
One of the grand challenges in physics and chemistry is to understand what actually happens during a chemical reaction. The nuclei in molecules move on the femtosecond (10-15 s) timescale, but the electrons in the molecules move on the attosecond (10-18 s) timescale. The R-matrix with time dependence (RMT) code is a leading code for the description of ultra-fast processes in general atoms and molecules. Scientists at Queen's University Belfast have been working on the RMT code, increasing its speed by up to a factor of 5 and reducing the amount of memory required by one or more orders of magnitude. Read more...
Understanding how bones develop and respond to disease and the use of implants
Scientists at the University of Hull have developed their simulation software to utilise ARCHER to model complete bones or large sections of bones. This offers the exciting opportunity to model skeletal development and adaptation. The potential benefits are enormous, ranging from a better understanding of both the fundamental biomechanics of bone and the cause and effects of musculoskeletal conditions, to better implant design. Read more...
TPLS: Optimised Parallel I/O and Visualisation
TPLS (Two Phase Level Set) is a powerful 3D Direct Numerical Simulation (DNS) solver that is able to simulate multi-phase flows at unprecedented detail, speed and accuracy with applications in energy (oil/gas pipeline flows, microelectronic cooling via phase-change), environment (carbon capture and cleaning) and health (flows in retinal capillaries). Scientists at the University of Edinburgh have been working on the code, converting the previously serial I/O to a scalable parallel implementation. This results in a halving of the time taken for each simulation from set-up to complete analysis. Read more...
Scalable and interoperable I/O for Fluidity
At Imperial College London, scientists have been working with the Fluidity CFD code to improve file I/O and the performance and scalability of the underlying PETSc library. The resulting performance increase is then utilised to improve the mesh initialisation performance of the Fluidity CFD code through run-time distribution and more efficient data migration. In addition to performance improvements the capabilities of PETSc's DMPlexDistribute interface have been extended to include load-balancing and re-distribution of parallel meshes, as well as the ability to generate multiple levels of partition overlap. Furthermore, support for additional mesh file formats has been added to DMPlex and Fluidity, including binary Gmsh, Fluent-Case. Read more...






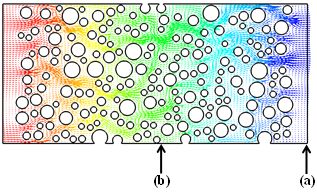



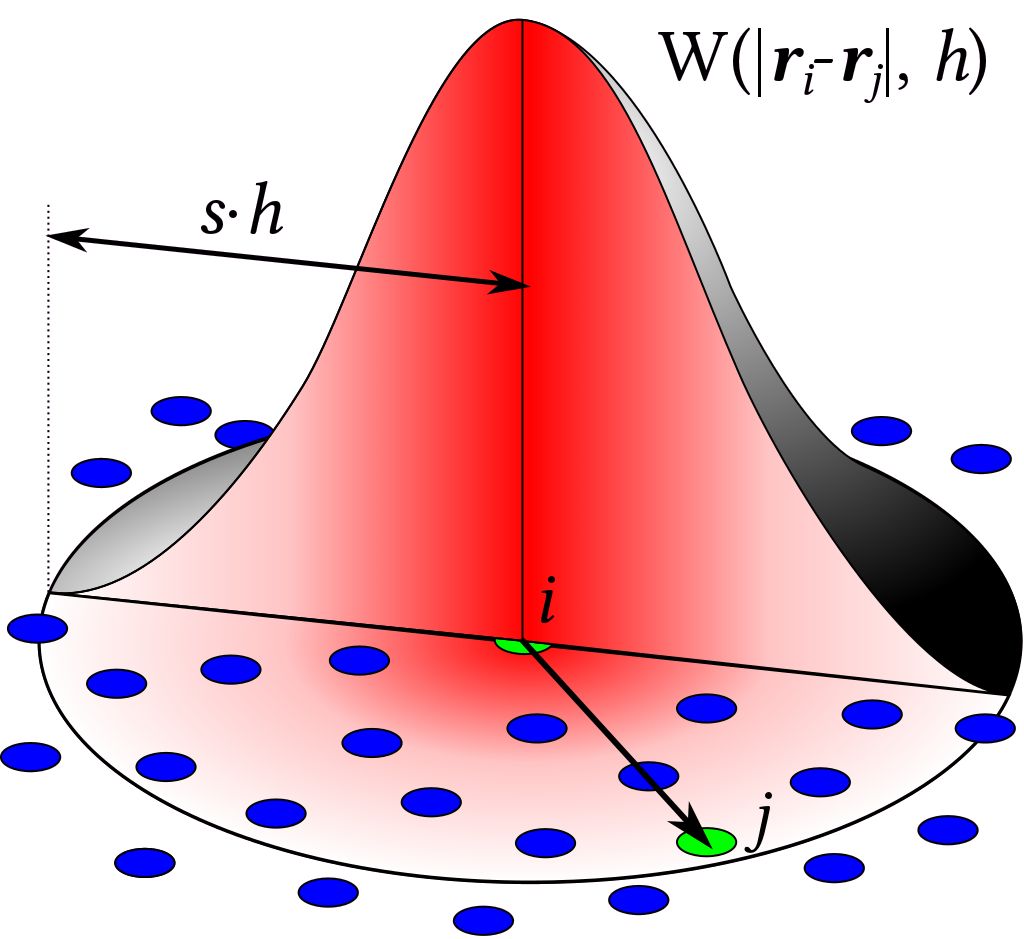
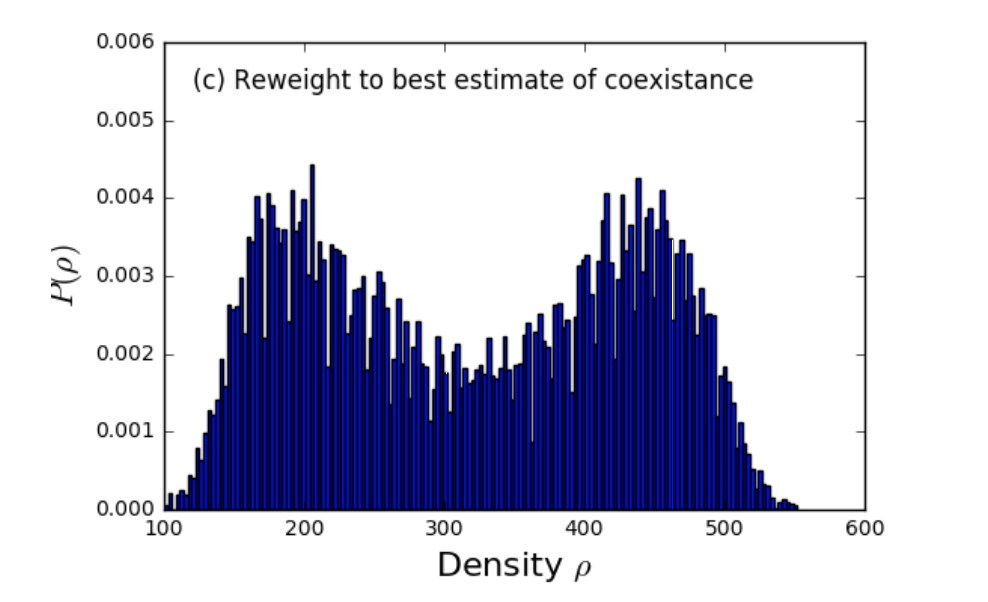

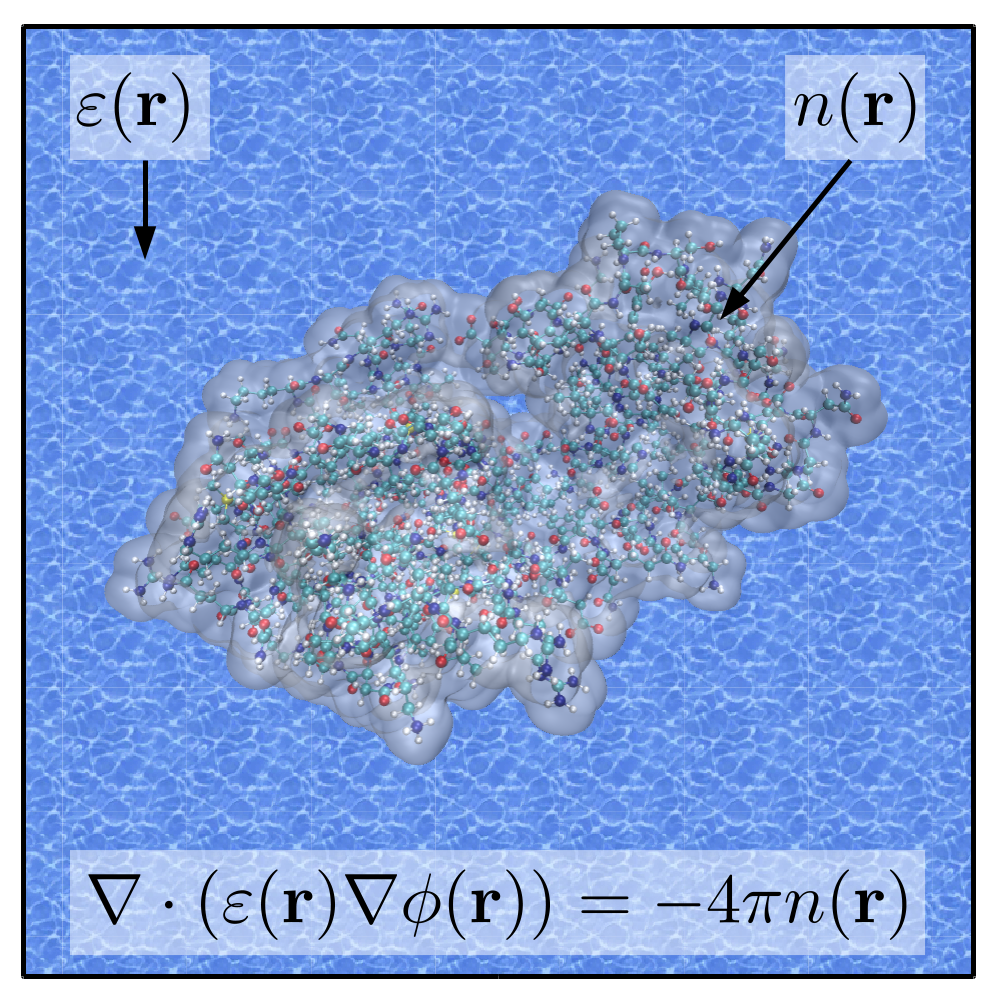







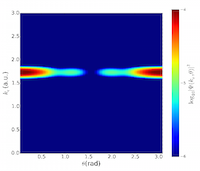





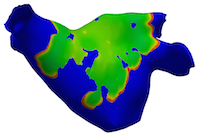
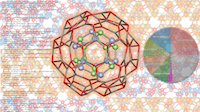
![Contour plot of the relative free energy density (summed enthalpy and entropy) for hydration of the cucurbit[7]uril system.](eCSE03-03/eCSE03-03-image-cropped.png)